Unit 4 Mutation
Pallister
Killian syndrome is a result of extra #12 chromosome material. There is usually
a mixture of cells (mosaicism), some with extra #12 material, and some that are
normal (46 chromosomes without the extra #12 material). Babies with this
syndrome have many problems. These include severe intellectual disability, poor
muscle tone, "coarse" facial features, and a prominent forehead. They
tend to have a very thin upper lip, with a thicker lower lip and a short nose.
Other health problems include seizures, poor feeding, stiff joints, cataracts
in adulthood, hearing loss, and heart defects. People with Pallister Killian
have a shortened lifespan, but may live into their 40s.
1. Para centric inversion: When centromere is not included in the inversion, it is called paracentric inversion.
It
involves a single break in chromosome. The broken piece gets attached to one
end of a non-homologous chromosome.
Chromosomal
Mutation: Deletion, Duplication, Inversion, Translocations,
Aneuploidy and polyploidy, Gene mutations Induced versus spontaneous mutations.
The
Chromosomes of every species possess a definite number of chromosomes. Each
chromosome has a specific structure. Which imparts, a specific phenotype. But
during the cell division due to certain accidents or irregularities the
alterations in chromosome number and its morphology takes place which changes
the phenotype. It changes the original structure and number of chromosomes.
Such type of change in genome involving chromosome parts or whole chromosome or
a set of chromosome is called as chromosomal mutations or chromosomal
aberrations.
Chromosomal
mutations:
I)
Structural changes in chromosomes
- Changes in number of genes i.e. a)
loss or deletion and b) addition or duplication
a)
Deletion: Loss – Changes in number of genes. Loss of portion of
chromosome is called deletion. Portions of the chromosomes without a centromere
lag in anaphase movement and are lost from reorganizing nuclei or digested by
nucleases. The chromosomes with deletions can never come back to normal
condition. If gametes arise from the cells having a deleted chromosome this
deletion is transmitted to the next generation.
The
deletion is the breakage of chromosome at any position, it can be terminal or
intercalary deletion.
Intercalary deletion:
Breakage
takes place at the middle of chromosome. It involves two breakages in the
chromosome.
Terminal Deletion:When
breakage takes place at the end of chromosome, there is only single breakage in
chromosome.
Example of deletions:
1)
Human babies missing a portion of
the short arm of chromosome 5 (outosome) have a distinctive cat like cry called
as “cri-du-chat” (cry of the cat) syndrome
2)
Turner syndrome: deletion of part
of the short arm of one X- chromosome.
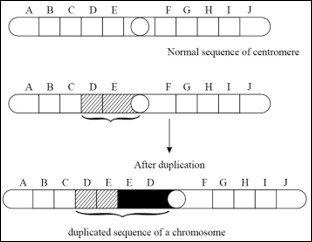
b) Duplications:
The
presence of a part of a chromosome in excess of the normal complement is known
as the duplication. Due to duplication some genes are present in more than two
doses. If duplication is present only on one of the two homologous chromosomes
during the meiosis the chromosome bearing the duplicated segment forms of loop
to maximize the pairing of homologous regions.
Types of duplications:
Tandem
duplication: In this type the duplicated region is situated just by the side of
the normal corresponding section of the chromosome and the sequences of genes
are the same in normal and duplicated region
2.
Reverse tandem duplication: In this type of duplication, the sequence of
genes in the duplicated region of a chromosome is just the reverse of normal
sequence.
3)
Displaced duplication:
Displaced
duplication heterobranchial on different arm.
4)
Transposed duplication: In this type of duplication, the
duplicated portion of chromosome becomes attached to different i.e.
non-homologous chromosome.
Examples of duplication:
Fig.
Transposed duplication
1)
Bar eye in drosophila: This phenotype
is characterized by narrower, oblong, bar shaped eye with few facets. It is due
to duplication of a segment of the X chromosome, called section 16A.
2)
In humans. Concerning the hemoglobin
gene, deletions result in lepore and Kenya variants of adult hemoglobin (Hba)
both causing anemia.
- Changes
in gene arrangement:
When
gene arrangement takes place Genes rotate within one chromosome or exchange of
parts of chromosomes takes place between chromosomes.
1) Inversion:
It
involves a rotation of a part of a chromosome or a set of genes by 1800
on its own axis. Here breakage and reunion of chromosomes takes place. Because
of this there is neither a gain nor a loss in genetic material but it is simply
a rearrangement of the gene sequence.
Inversion
is of following two types.
The
location of the centromere relative to inverted segment determines the genetic
behavior of the chromosomes.
It
is of two types.
1. Para centric inversion: When centromere is not included in the inversion, it is called paracentric inversion.
2) Pericentric
inversion: When inversion involves centromere it is called
pericentric inversion
Advantages of inversions :
Fertility of inversion homozygotes and sterility of inversion heterozygotes
leads to establishment of two group (or varieties) which are mutually fertile
but do not breed well with the rest of the species.
2)
Translocations:
The
transfer of a part of chromosome or a set of genes to a non-homologous
chromosome is called as translocation.
The
rearrangement of genes takes place The sequence and position of gene changes.
There is no addition or loss of gene during translocations.
It
is of following types :
a)
Simple translocations:
b)
Shift translocation:
In
this type of translocation, the broken segment of one chromosome get inserted
interstitially in a non-homologous chromosome.
c)
Reciprocal translocation:In this type, a segment from one chromosome is get
exchanged with a segment from another non-homologous chromosome, In this way
two translocation chromosomes are simultaneously achieved.
Example:
Philadelphia
chromosome:
A
translocation between the long arms of chromosome g and 22 often in the white
blood cells of patients with chronic myeloid leukemia
II)
Changes in number of chromosomes: It includes;
1. Loss
or gain of the part of the chromosome set (aneuploidy)
2. Loss
or gain of whole chromosome set (euploidy)
The
phenomenon of variations in the number of chromosomes is called heteroploidy.
It is of two types, euploidy and aneuploidy.
A)
Euploidy
In
this condition, an organism either loses a complete set of chromosomes or
acquires one or more additional sets of chromosomes over and above the two sets
of diploid complement. On this basis, euploidy can be categorized into two
types – monoploidy (haploidy) and polyploidy.
a)
Monoploidy or haploidy
Monoploidy
have only one set of chromosomes. Haploids have half the somatic chromosome
number. In diploid organisms, monoploids and haploids are identical while they
differ in tetra or hexaploid organisms.
In
certain organisms (e.g. honeybee) haploid offspring are produced as a routine
which arise parthenogenetically from the unfertilized eggs. Haploids can be
artificially induced by many methods like X-ray treatment, delayed pollination,
culturing pollen grain and temperature shocks.
Haploids
are smaller in size as compared to diplods. Their leaves, flowers, fruit and
seeds are comparatively small. They are also less resistant and comparatively
weak. The haploids have just univalent without any homologous chromosomes to
pair with during meiosis. These univalents are distributed at random during
anaphase-I of meiosis producing monosomatics. A cell with the haploid number of
8, for example, may produce at the first meiotic division two daughter cells
with chromosome numbers anywhere from 0 to 8. That is why the haploids are
mostly sterile. The haploids which take part in reproduction ( e.g. male
honeybee) form their gametes through mitosis and not meiosis.
b)
Polyploidy
It
is the phenomenon of having more than two sets of chromosomes. It occurs due to
failure of chromosomes to separate at the time of meiotic anaphase either due to nondisjunction
or due to nonformation of spindle. Polyploidy can also be artificially induced
by application of colchicines and granosan. Depending upon the number of genomes
present in polyploidy, an organism may be triploid (3n), tetraploid (4n),
pentaploid (5n), hexaploid (6n) and so on. Polyploids with odd number of
genomes (i.e. triploids, pentaploid) are sexually sterile because the
odd chromosomes do not form synapsis during meiosis. They are, therefore propagated
vegetatively, e. g. Banana, Pineapple. Polyplods also do not cross breed freely
with diploids. The polyploidy occurs much more frequently in plants than in
animals. It is so because tha plants can propogate vegetatively. Thus the
sterile triploids, pentaploids etc. can be maintained from generation to
generation. Animals mostly reproduce sexually through gamete formation. As
already mentioned gamete formation is not normal in ployploids due to failure
of chromosome pairing during meiosis. The ployploids can be maintained and are
seen in animals, which reproduce exclusively by diploid parthenogenesis (some
crustaceans). The poliploids usually show gigas effect at both
morphological and biochemical levels due to increase in frequency of dominant
alleles. The gigas effect results in
larger size and higher yield. Some groups of organisms, primarily plants have
many ployploid members. An estimated 30 to 80 % of all flowering plant species
are ployploids as are 95% of ferns. Plyploidy is rare in gymnosperms and fungi.
Polyploidy
is of three types – autopolyploidy, allopolyploidy and autoallopolyploidy.
i)
Autopolyploidy – In
this case there is a numerical increase in the number of the same genome, e.g.
autotriplod (AAA), autotetraploid (AAAA) in case of a cell having AA genome.
Some of the crop and garden plants are autopolyploids, e.g. Maize, Rice,
Gram, Lawn grass (Cynodon), Grapes, Watermelons, Sugar beet, Potato.
Autoploid are detected by multivalent formation during meiosis. Because of no
seed formation ( in autoploids with odd number of sets) or poor seed formation,
autoploids are preferred in plants in which seeds are of no economic
importance. Sugar beet and water melon and lawn grass are mainly triplods and
potato is mainly tetraploid. Among autoploids, tetraploids are meioticallyt
more stable. Autoployploidy induces gigas effect.
ii)
Allopolyploidy - It
has developed through hybridization between two species folloed by doubling of
chromosomes (e.g. AABB) Allotetraploid is the common type. For example,
an allotetraploid between two species A and B will be formed if F1 hybrid (AB)
of these two species undergoes chromosomal doubling or produces diploid
gametes. In most of the cases the hybrids between the two species are sterile
because of failure of pairing between unrelated chromosomes at the time of
meiosis. Although very rare, but still it is possible that both the sets of
chromosomes of the hybrid enter one gamete, producing diploid gametes. Such
diploid gametes on fertilization produce allotetraploids. Sometimes the zygote
(which has been formed by hybridization between two species) undergoes
chromosomes doubling but fails to undergo forst mitotic division. This event
will also produce an allotetraploid. Allotetraploids
like AABB are also called amphidiploids. Alloployploids function as new
species, e.g. Wheat, American cotton, Nicotiana tabacum. Two recently
produed allopolyploids are Raphanobrassica and Triticale.
Allopolyploids are more advantageous as besides possessing traits of different
species and benefits of ployploiy, they are meiotically also stable.
iii) Autoalloployploidy – In
which one genome is in more than diplod state. Commonly autopolyploids are
hexaploids (AAAABB), e.g. Helianthustuberosus
B)
Aneuploidy
It
is a condition of having fewer or extra chromosomes than the normal diplod
chromosome complement of an organism. It is of two types, hypoploidy or loss of
chromosomes and hyperploidy or addition of chromosomes. The organisms showing
aneuploidy are known as aneuploids. They are represented by yhe number of
affected chromosomes with the suffix-somic, e. g. nullisomic, monosomic,
trisomic etc. Aneuploidy usually arises due to nondisjunction of the two
chromosomes of homologous pair so that one gamete comes to have an extra
chromosome (N+1) while the other becomes deficient in one chromosome (N-1).
Fusion with similar or normal gametes will produce four types of aneuploids.
N
x (N-1) = 2N-1
(N-1)
x (N-1) = 2N-2
N
x (N+1) = 2N+1
(N+1)
x (N+1) = 2N=2
Another
way of formation of aneuploid is through loss of chromosomes from a normal or
ployploid karyotype due to faulty mitosis. Aneuploidy can be further divided
into 2 types
I)
Hyperploidy - When there is addition of either a single
chromosome i. e. trisomy (2n + 1) or a
pair of chromosome (2n + 2) called tetrasomy.
a)
Trisomic (2N+1): In
this case there is one chromosome in triplicate. Double trisomic has two
different chromosomes in treiplicate (2N+1+1). Trisomics show a number of
changes some of which are lethal. Down’s syndrome is an example of trisomy
where chromosome number 21 is in triplicate. Trisomy is detected at the time of
meiosis by formation of trivalents. Trisomy of single chromosome will show one
trivalent while double trisomic will show two trivalents. In man, the example
of trisomy are Down’s syndrome (Trisomy- 21), Edward syndrome (Trisomy- 18) and
Patau syndrome (Trisomy- 13).
b) Tetrasomic –
In this case one chromosome is represented four times. Tetrasomics show more
variability than trisomics and tetrasomicsare believed to have given rise to
new species through secondary polyploidy, e.g. Apple, Pear. The general
chromosome formula for tetrasomics is 2n+2 rather than 2n+1+1. The formula
2n+1+1 represents a double trisomic. Tetrasomics form quadrivalent at the time
of meiosis.
II)
Hypoploidy:
It occurs mainly due to subtraction or loss of a single chromosome, called
monosomy (2n – 1) or due to loss of one
pair of chromosome called Nullisomy (2n-2).
a)
Monosomic (2N-1)-
In this case one chromosomes lacks its homologue. Its general formula is 2n-1.
In case one chromosome of some other pair is also lost, it is known as double
monosomic and is represented as 2N-1-1. Monosomic is generally weaker than the
normal form. Turner’s syndrome is an example of sex monosomic in human being
(44+X).
b)
Nullisomic (2N-2)
– It is deficient in a complete pair of homologous chromosomes. Nullisomics do
not survive except among polyploids. At the time of meiosis, nullisomics will
have one bivalent less.
III)
They
are aneuploid with both hypoploidy and hyperploidy, e.g., 2N+1A-1B
Gene
Mutation
These
mutations are caused by a change in the nucleotide type and sequence of a DNA
segment representing a gene. The first recorded gene mutations are Ancon Sheep
and hornless (polled) cattle.ofThe first scientific study of gene mutation
started with the discovery of white eye trait in Drosophilia by Morgan.
All genes have potential to undergo mutations but it differs from gene to gene.
Mutation can occur in every conceivable direction and to every conceivable
degree. Mutations can occur in both somatic and germinal cells. Mutations may
be lethal, harmful, neutral or advantageous. Most of the mutations are
recessive and harmful. Mutator genes increase the rate of mutation in some
genes while antimutator genes reduce the frequency of mutation of certain
genes.
Reverse
mutations
Most
mutant events consist of a change from wild or normal type to a new mutant
genotype. Such mutation events are known as forward mutations. In contrast to
the back or reverse mutations, in which athe mutant genotype changes back to
the wild type.
Spontaneous
verse Induced mutations
1)
Spontaneous mutations–They occur randomly and automatically in
nature. The possible reasons are:
i)
Background radiations -
They are present in natural surrounding and come from various sources, e.g.,
sun, radioactive minerals.
ii)
Tautomers –
All the four nitrogen bases also occur in their tautomeric or isomeric states,
forming either imino group (-COH, e.g., thymine, guanine) instead of
ketogroup (=CO). Tautomers pair with different bases so as to cause a change in
the sequence like AT to CG.
iii)
Deamination of Cytosine –
Cytosine slowly deaminates to produce uracil which pairs with adenineresulting
in change in base pairing and thus causing mutation.
iv)
Copy error –
Many steps are involved in replication, transcription and translation. Any
wrong choice of entry of different group wll results in mutation. Most of these
errors are rectified during proof reading but a few escape this rectification
and thus will cause mutations.
2)
Induced mutations – These mutations are produced artificially
with the help of certain agents, mutagens. Any extracellular physical or
chemical factor that has the ability to cause mutations or increases the
frequency of mutations is called mutagen. Following are some mutagenic agents
that cause mutations:
I)
Physical mutagens – They are of two types, temperature and
high energy radiations.
i)
Temperature: Rise
in temperature increases the rate of mutations. Increased temperature breaks
the hydrogen bonds between the two strands of DNA and denatures it. It disturbs
the synthetic process connected with replication and transcription. In Rice,
low temperature increases the rate of mutations.
ii)
High energy radiations
– The biological effect of different radiations is not equally harmful. The
harmful effect is determined by the penetration and ionizing power of the rays.
Radiations are broadly divided into two categories:
a)
Ionizing radiations
– These include X-rays, gamma rays, alpha rays, beta rays, neutrons and
protons. Alpha and beta rays do not penetrate beyond the human skin and
therefore, usually do not affect internal body cells. The gamma rays and X-rays
collide with the biomolecules at high speed and eject electrons from the outer
shells of atoms. These atoms after losing electrons become positively charged
ions. The ejected electrons after losing their energy get attached to other
atoms which then become negatively charged ions. These ions undergo chemical
reactions to neutralize their charge to reach a stable state. During these
reactions, the mutagenic effects of ionizing radiations are produced. The
ionizing radiations produce breaks in the chromosomes. These breaks then lead
to loss of chromosomes, chromosome segments, deficiencies, duplications,
translocations or inversions.
X-rays are known to
deaminate and dehydroxylate nitrogen bases, form peroxides and oxidize
deoxyribose. Muller was the first to induce mutations in Drosophila with the
help of the X-rays.
b)
Non-Ionizing radiations – These
radiations which include ultraviolet rays, have longer wave lengths and carry
much lower energy. Their penetration power is, therefore, much less than the
ionizing radiations. In humanbeings, UV rays are usally absorbed by the skin
and the gonads remain unaffected.
The UV rays may be
absorbed by the nucleic acids. Two adjacent pyrimidines of the same DNA strand
form covalent bonds forming dimmers. The thymine dimer is formed most
frequently. Dimerization interferes with the proper base pairing of thymine
with adenine and may result in the pairingof thymine with guanine.
II)
Chemical mutagens: Chemical mutagen is a mutation agent which
is in the form of chemical substance, it can mimic nitrogen base in normal DNA,
but they cannot couple during DNA replication. Moreover, chemical mutagen has
an ability to insert between nitrogen bases and disturb DNA replication. They
are of several types. The common ones are alkylating agents, base analogues and
acridines.
i)
Alkylating Agents: This
is the most powerful group of mutagens. They induce mutations especially
transitions and transversions by adding an alkyl group (either ethyl or methyl)
at various positions in DNA. Alkylation produces mutation by changing hydrogen
bonding in various ways. The alkylating agents include ethyl methane sulphonate
(EMS), methyl methane sulphonate (MMS), ethylene imines (EI), sulphur mustard,
nitrogen mustard, etc.
ii) BaseAnalogues: Base
analogues refer to chemical compounds which are very similar to DNA bases. Such
chemicals sometimes are incorporated in DNA in place of normal base during
replication. Thus, they can cause mutation by wrong base pairing. An incorrect
base pairing results in transitions or transversions after DNA replication. The
most commonly used base analogues are 5 bromo uracil (5BU) and 2 amino purine
(2AP).
iii) Acridine Dyes: Acridine
dyes are very effective mutagens. Acridine dyes include, pro-flavin, acridine orange,
acridine yellow, acriflavin and ethidium bromide. Out of these, pro-flavin and
acriflavin are in common use for induction of mutation. Acridine dyes get
inserted between two base pairs of DNA and lead to addition or deletion of
single or few base pairs when DNA replicates. Thus, they cause frameshift
mutations and for this reason acridine dyes are also known as frameshift
mutagens.











No comments:
Post a Comment